Integration of scATAC data with scPoli
In this notebook we demonstrate an example workflow of scATAC data integration. We integrate data obtained from the .NeurIPS 2021 multimodal single cell data integration. The data can be downloaded from GEO.
[1]:
import numpy as np
import scanpy as sc
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from scarches.models.scpoli import scPoli
import warnings
warnings.filterwarnings('ignore')
%load_ext autoreload
%autoreload 2
WARNING:root:In order to use the mouse gastrulation seqFISH datsets, please install squidpy (see https://github.com/scverse/squidpy).
INFO:pytorch_lightning.utilities.seed:Global seed set to 0
/home/icb/carlo.dedonno/anaconda3/envs/scarches/lib/python3.10/site-packages/pytorch_lightning/utilities/warnings.py:53: LightningDeprecationWarning: pytorch_lightning.utilities.warnings.rank_zero_deprecation has been deprecated in v1.6 and will be removed in v1.8. Use the equivalent function from the pytorch_lightning.utilities.rank_zero module instead.
new_rank_zero_deprecation(
/home/icb/carlo.dedonno/anaconda3/envs/scarches/lib/python3.10/site-packages/pytorch_lightning/utilities/warnings.py:58: LightningDeprecationWarning: The `pytorch_lightning.loggers.base.rank_zero_experiment` is deprecated in v1.7 and will be removed in v1.9. Please use `pytorch_lightning.loggers.logger.rank_zero_experiment` instead.
return new_rank_zero_deprecation(*args, **kwargs)
WARNING:root:In order to use sagenet models, please install pytorch geometric (see https://pytorch-geometric.readthedocs.io) and
captum (see https://github.com/pytorch/captum).
WARNING:root:mvTCR is not installed. To use mvTCR models, please install it first using "pip install mvtcr"
WARNING:root:multigrate is not installed. To use multigrate models, please install it first using "pip install multigrate".
[2]:
sc.settings.set_figure_params(dpi=100, frameon=False)
sc.set_figure_params(dpi=100)
sc.set_figure_params(figsize=(3, 3))
plt.rcParams['figure.dpi'] = 100
plt.rcParams['figure.figsize'] = (3, 3)
[3]:
adata = sc.read('../datasets/GSE194122_openproblems_neurips2021_multiome_BMMC_processed.h5ad')
# remove those that appear in fewer than 5% of the cells
sc.pp.filter_genes(adata, min_cells=int(adata.shape[0] * 0.05))
We select the ATAC features, specify the covariates we want to use as condition and cell type annotation and transform the data from reads to fragments.
[4]:
adata = adata[:, adata.var['feature_types']=='ATAC']
adata.X = adata.X.todense()
adata.X = adata.X.astype('float32')
[5]:
condition_key = 'Samplename'
cell_type_key = 'cell_type'
[7]:
from scarches.models.scpoli._utils import reads_to_fragments
adata_fragments = reads_to_fragments(adata, copy=True)
We instantiate a model with a Poisson likelihood and train it.
[10]:
scpoli_model = scPoli(
adata=adata_fragments,
condition_keys=condition_key,
cell_type_keys=cell_type_key,
hidden_layer_sizes=[100],
latent_dim=25,
embedding_dims=5,
recon_loss='poisson',
)
scpoli_model.train(
n_epochs=100,
pretraining_epochs=95,
use_early_stopping=False,
alpha_epoch_anneal=1000,
eta=0.5,
)
Embedding dictionary:
Num conditions: [13]
Embedding dim: [5]
Encoder Architecture:
Input Layer in, out and cond: 16134 100 5
Mean/Var Layer in/out: 100 25
Decoder Architecture:
First Layer in, out and cond: 25 100 5
Output Layer in/out: 100 16134
Initializing dataloaders
Starting training
|████████████████████| 100.0% - val_loss: 21214.68 - val_cvae_loss: 21208.48 - val_prototype_loss: 6.20 - val_labeled_loss: 12.40
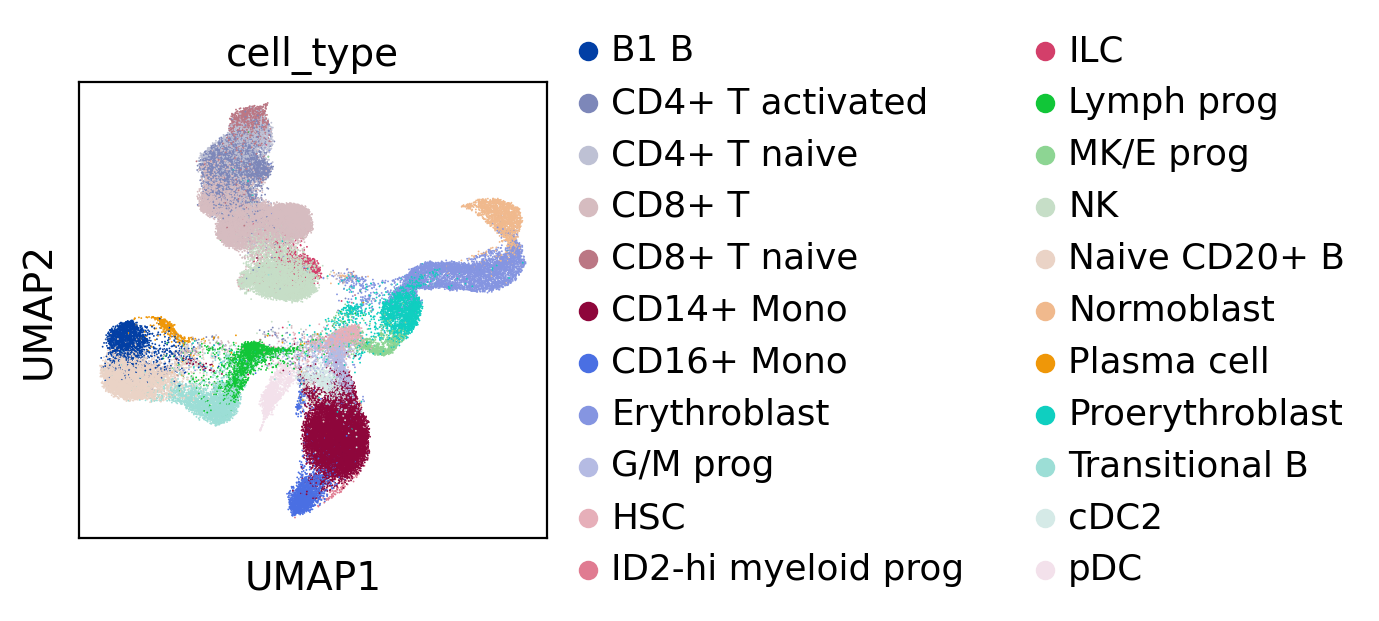
Plotting cell embeddings
[11]:
#get latent representation of reference data
scpoli_model.model.eval()
data_latent = scpoli_model.get_latent(
adata_fragments,
mean=True
)
adata_latent = sc.AnnData(data_latent)
adata_latent.obs = adata_fragments.obs.copy()
sc.pp.pca(adata_latent)
sc.pp.neighbors(adata_latent)
sc.tl.umap(adata_latent)
OMP: Info #276: omp_set_nested routine deprecated, please use omp_set_max_active_levels instead.
[12]:
sc.pl.umap(adata_latent, color='cell_type')
[23]:
adata_emb = scpoli_model.get_conditional_embeddings()
adata_emb.obs = adata.obs.groupby('Samplename').first().reindex(adata_emb.obs.index)
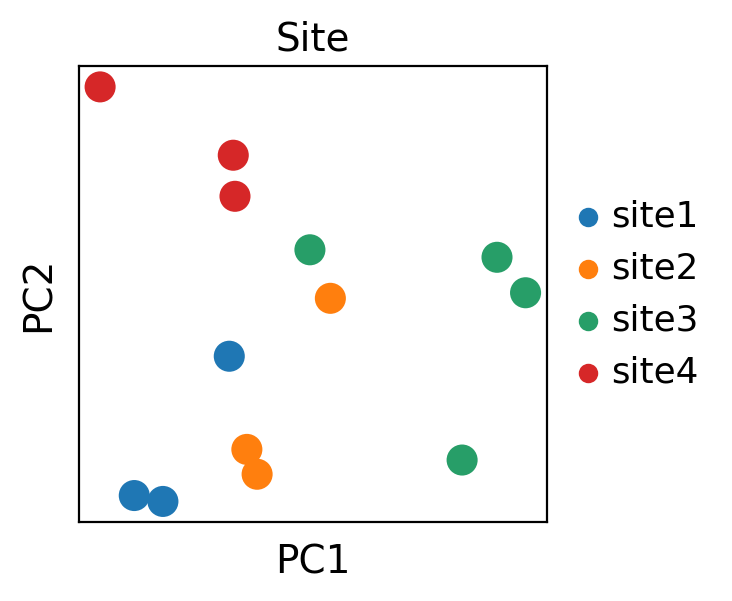
Plotting sample embeddings
[24]:
sc.pp.pca(adata_emb)
sc.pl.pca(adata_emb, color='Site', size=500)